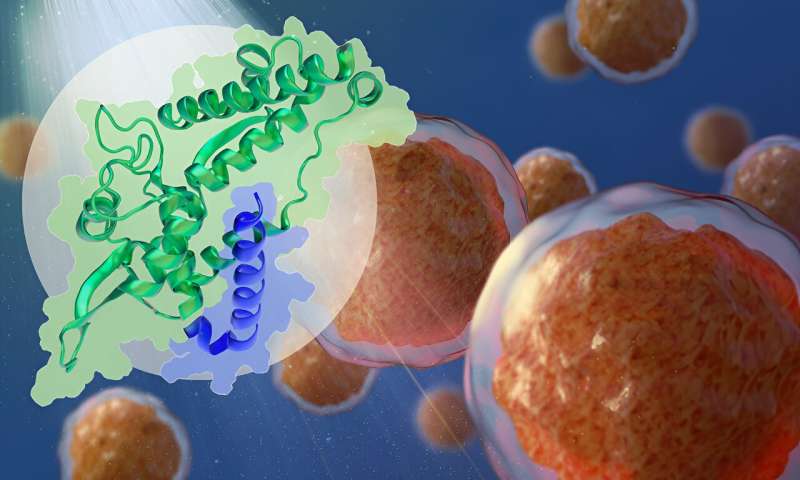
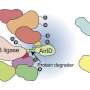
The new study demonstrates the structural basis of the interaction between cancer-promoting proteins MAGEA4 (green) and RAD18 (blue). Credit: Isabel Romero Calvo/EMBL
Researchers from the Bhogaraju Group at EMBL Grenoble have gained new insights into how a cancer-relevant family of proteins bind their targets. The results of the study, published in The EMBO Journal, could potentially help in the development of drugs against certain chemotherapy- and radiotherapy-resistant cancers.
The melanoma antigen gene (MAGE) family consists of more than 40 proteins in humans, most of which are only present in the tests under healthy conditions. However, in many cancers, these proteins are found in high levels in tissues where they are not usually expressed and are believed to play a role in promoting cancer progression.
One such MAGE protein—MAGEA4—is known to interact with RAD18, a protein known to be abundant in some cancer cells. The latter is part of the molecular machinery that helps the cell repair damage to its DNA. High levels of RAD18 are responsible for the resistance of several cancers to genotoxic (DNA-damaging) chemotherapy or radiotherapy.
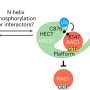
RAD18 functions by attaching little molecular tags—called ubiquitin—to different proteins. This tag, like a postal stamp, tells the cell what the fate of that protein would be. RAD18 can also attach this tag to itself—a process called autoubiquitination. This targets it for degradation, i.e., tells the cell to get rid of excessive levels of this protein.
The Bhogaraju Group at EMBL Grenoble uses structural and cell biology–based approaches to study such ubiquitin-based pathways in normal physiology and disease. The team, in collaboration with the Hennig Group at EMBL Heidelberg, decided to look more deeply into the interaction between the proteins MAGEA4 and RAD18 using AlphaFold, an artificial-intelligence based tool that allows scientists to predict the structure of proteins.
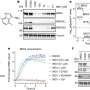
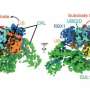
The team, which included Bhogaraju Group Ph.D. student Simonne Griffith-Jones and postdoc Urbi Mukhopadhyay, found that MAGEA4 has a groove that can bind a section of the RAD18 protein, which prevents the latter from attaching ubiquitin groups to itself and subsequently getting degraded.
Interestingly, the researchers could use a short synthetic protein fragment, mimicking the part of RAD18 that binds the groove in MAGEA4, to block the interaction between the two proteins. This could potentially pave the way for the design of drugs that target this complex and prevent RAD18 build-up within cancer cells.
The researchers also found a very similar groove in another MAGE family protein, which is used to regulate a different cancer-promoting protein. They believe this groove may be a general feature of the MAGE family, used to mediate binding to cancer-relevant proteins.
In addition to the groove, the scientists also observed that two parts within the RAD18 protein interact with each other, which helps it attach the ubiquitin tag to a protein which promotes cancer cell survival. They could block this function using genetic strategies, implying that future drugs designed to block this interaction could potentially re-sensitize cancer cells which have gained resistance to chemotherapies or radiotherapies.
"We are excited with these data because our findings seem to be applicable to many MAGEs opening a gateway into targeting MAGEs which drive cancers," said Sagar Bhogaraju, Group Leader at EMBL Grenoble. "We are currently working on developing methods to screen compounds that bind the newly discovered hotspot of MAGEs."
More information: Simonne Griffith-Jones et al, Structural basis for RAD18 regulation by MAGEA4 and its implications for RING ubiquitin ligase binding by MAGE family proteins, The EMBO Journal (2024). DOI: 10.1038/s44318-024-00058-9
Journal information: EMBO Journal
Provided by European Molecular Biology Laboratory