This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Ubiquitin research provides new insights into the cell's labeling machine
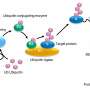
Ubiquitin ligases regulate many processes in living cells. Depending on how many ubiquitin molecules they attach to the target proteins and in which way, they control the protein's fate.
"The ubiquitin code can direct target proteins to specific cellular sites of action, activate them, trigger their incorporation into protein complexes or their degradation," explains Sonja Lorenz, head of the Ubiquitin Signaling Specificity research group at the MPI. It is thus crucial that ubiquitin ligases reliably recognize the corresponding proteins among tens of thousands of molecules and transmit the correct code.
How this molecular recognition works is a focus of Lorenz and her team's research. "We want to uncover the structural principles of this specific recognition and reveal how they enable different ubiquitin ligases to control specific processes," the biochemist says. This knowledge is not only important for understanding the role of ubiquitin ligases in normal or pathologically altered processes in living cells. It also helps to make the ubiquitin system more accessible for therapeutic applications.
Dynamic ubiquitin system
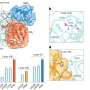
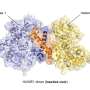
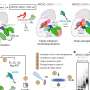
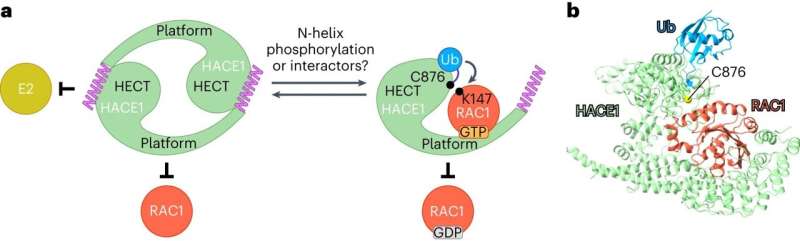
However, it is difficult to observe ubiquitin ligases bound to their target proteins. "The ubiquitin system is designed to be highly dynamic," Lorenz says. "The underlying interactions are therefore very weak and of short duration." Nevertheless, the group has succeeded in visualizing such an interaction between the ubiquitin ligase HACE1 and its primary target protein, the signaling mediator RAC1.
The researchers used a chemical trick to do this. "With the help of chemical crosslinking, we could stabilize the complex at just the right time to make it accessible for structural studies using cryo-electron microscopy," says the research group leader. In collaboration with the Facility for Cryo-Electron Microscopy at the MPI, headed by Christian Dienemann, the team succeeded, for the first time, in obtaining a "snapshot" of the full-length HACE1-RAC1 complex.
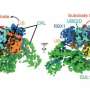
Through additional biophysical and functional experiments, Lorenz's team discovered that the particular architecture of the ubiquitin ligase ensures that HACE1 and its target protein interact with specificity. Their studies show that the ubiquitin ligase forms a platform on which the target protein is positioned. This enables the ligase to reliably recognize its target protein and distinguish it from other proteins. HACE1 can also determine whether RAC1 is in an active or inactive state. This is crucial because RAC1 ought to be tagged with ubiquitin only in its active form.
However, the ubiquitin ligase HACE1 itself can also adopt an active or inactive state, report Jonas Düring and Madita Wolter, the joint first authors of the paper now published in Nature Structural & Molecular Biology. "Two HACE1 molecules can bind to each other to form a complex, a so-called dimer, and thus switch themselves off," explains Düring. Wolter adds, "HACE1 is only active as a single molecule. Dimerization is, therefore, an important regulatory mechanism."
"If HACE1 no longer functions correctly in the cell or is missing altogether, this can lead, for example, to developmental perturbations of the nerves that are associated with human diseases," explains Lorenz.
"Owing to the insight into the precise interaction of HACE1 and its target protein RAC1, we will be able to better understand how genetic changes in this molecular labeling machine can disrupt target protein recognition." The new findings also contribute to the understanding of related ubiquitin ligases, whose target recognition mechanisms are still poorly understood.
More information: Jonas Düring et al, Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1, Nature Structural & Molecular Biology (2024). DOI: 10.1038/s41594-023-01203-4
Journal information: Nature Structural & Molecular Biology
Provided by Max Planck Society