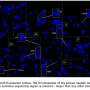
Chromosome satellite for both species (Chinese chestnut on the left, American chestnut on the right). Like all chromosomes, these are made of chromatin—a mix of DNA molecules and proteins. There are several types of chromatin. The bright blue tip in the Chinese chestnut indicates heterochromatic DNA, and the lighter purple color on the tip may represent the euchromatic DNA. The entire region of the American chestnut satellite appears to be euchromatic. Credit: USDA Forest Service image by Nurul Faridi.
The chromosomes of American and Chinese chestnut are not so similar after all, at least in one key region of the genome—the nucleolus organizing region (NOR).
The finding, published in Scientific Reports, has major implications for anyone with the goal of conferring blight resistance to American chestnuts through hybridization with the Chinese chestnut.
"This is an unprecedented finding in the field of plant cytology," says Nurul Faridi, a Forest Service geneticist and lead author of the study.
Traditional backcross breeding, involving hybridization between two species, aims to combine an ideal mix of traits from two species without genetic engineering. Backcross breeding can only succeed when the chromosomes of both species are compatible. Because Chinese-American chestnut hybrids are viable, people have assumed that the two species are highly compatible. But the new study reveals significant differences in the NOR of the two species.
The NOR is part of every plant and animal cell. It carries the genetic instructions for making ribosomes—the molecular machines making proteins essential for life.
The NOR is located near the end of the short arm of a particular chromosome. It is present in both species, but in Chinese chestnut, it is packed with a type of DNA known as heterochromatin and constitutes about 25% of the chromosome.
The structure and composition of this DNA surprised the researchers—it is highly condensed, lacking gene content, and transcriptionally inactive. In contrast, the American chestnut satellite is very small and appears to be euchromatic. Euchromatic regions of DNA are transcriptionally active.
Faridi first noticed a small pair of Chinese chestnut chromosomes exhibiting very bright fluorescence with a specialized microscope, a UV filter, and a dye that binds to the DNA.
Faridi used fluorescent in situ hybridization (FISH) to further analyze the discovery.
"Our high-quality FISH images provide unequivocal evidence of this unique DNA arrangement," says Faridi. "These images are not just pictures; they are a testament to the dynamic nature of genetic material."
Faridi has been working with FISH since 1991 and has extensive experience preparing plant chromosomes for analyses. Well-separated chromosomes from enzymatically digested root tips that are mostly free of cell walls, nuclear membranes, and cytoplasmic debris are best for FISH.
Most FISH imagery is obtained from animal cells, as plant cells, and especially trees, are more challenging to work with. Faridi has found that chestnuts are far more difficult to work with than pine and poplar.
The researchers will use a technique called oligonucleotide FISH for further investigation. Oligo-FISH uses short specific DNA probes acquired from DNA sequencing. Since the entire genomes of American and Chinese chestnuts have been sequenced, oligo-FISH will allow the researchers to conduct detailed genetic studies to discern subtle genomic differences. The technique is especially useful for studying hybrids since it can indicate which parent a gene is from.
The progress in developing American chestnut hybrids with the American chestnut's height and the Chinese chestnut's blight resistance has been significant. However, the most advanced hybrids do not currently have enough blight resistance for restoration, as previous Forest Service research has shown.
More information: Nurul Faridi et al, Cyto‑molecular characterization of rDNA and chromatin composition in the NOR‑associated satellite in Chestnut (Castanea spp.), Scientific Reports (2024). DOI: 10.1038/s41598-023-45879-6
Journal information: Scientific Reports
Provided by USDA Forest Service