This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Researchers identify 'unicorn' defense mechanism that protects bacteria from antibiotics
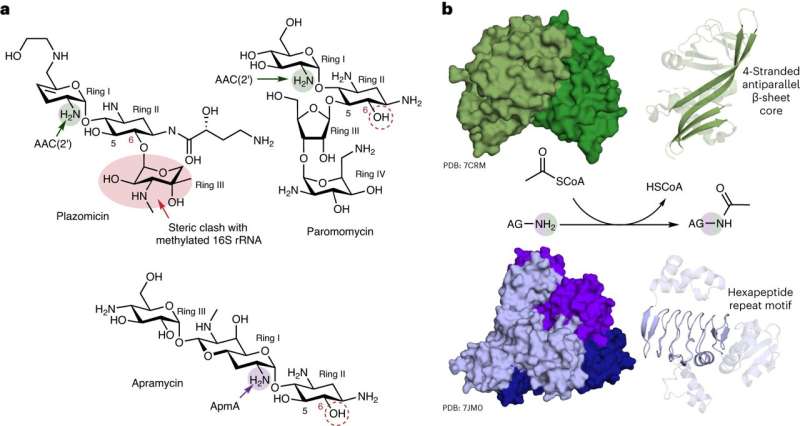
Researchers at McMaster University have discovered unique characteristics of a mechanism used by bacteria to resist an important class of antibiotics. The new research, published in Nature Chemical Biology, shows that resistance to aminoglycoside drugs—used to treat a variety of infections—is far more complex than initially thought.
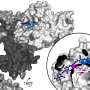
Lead investigator Gerry Wright, professor of Biochemistry and Biomedical Sciences at McMaster, says his lab observed never-before-seen versatility in ApmA, a long-studied bacterial resistance gene. The research showed that the gene can uncharacteristically enable bacteria to perform different functions against different antibiotics.
Of the hundred-or-more aminoglycoside resistance enzymes known to researchers, Wright says only this one has exhibited such nimble behavior.
"It's a unicorn," he says. "It looks different, it operates differently, and it belongs to an entirely different family of enzymes. It's completely different from all of the resistance mechanisms that we associate with this class of antibiotic."
Wright, a member of the Michael G. DeGroote Institute for Infectious Disease Research, says aminoglycosides were among the earliest antibiotics with clinical relevance—and the first-ever to be useful against tuberculosis. But because they've been prescribed since the 1940s, he says "resistance to them has become a real issue"—except in the case of apramycin.
"The antibiotic apramycin avoids most mechanisms of resistance, and so it is a strong candidate for new clinical applications," he says. "Unfortunately, this mechanism that we've been studying is not one that the drug can avoid."
Wright says his lab's recent discovery is significant because apramycin is currently in clinical trials and, should it pass through, having a thorough understanding of how bacteria might resist the drug will be crucial to extending its utility.
"If we're going to bring this drug to market, then we['d] better know what the enemy is," he says. "Learning more about this unique resistance mechanism could inform follow-on research into next-generation apramycin or diagnostics that could detect ApmA in bacteria."
More information: Emily Bordeleau et al, Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance, Nature Chemical Biology (2023). DOI: 10.1038/s41589-023-01483-3
Journal information: Nature Chemical Biology
Provided by McMaster University