This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Exploring the genetic potential of eggplant's wild relatives for sustainable agriculture
In the pursuit of sustainable agriculture, enhancing nitrogen use efficiency (NUE) in crops stands as a primary objective. With the prolific use of nitrogen (N) fertilizers since the 20th century, agricultural productivity has seen remarkable growth. However, excessive use of N fertilizers has resulted in serious environmental threats and energy consumption.
Crop wild relatives (CWR) provide valuable genetic resources to address this issue through breeding programs. Wild relatives of eggplant (Solanum melongena L.) are classified into primary (GP1), secondary (GP2), and tertiary (GP3) gene pools, which are unexploited gene pool. Yet, direct utilization of CWRs in breeding is complex due to inherent genetic barriers. This underscores the imperative to develop and study advanced backcrosses (ABs) for seamlessly incorporating these beneficial traits.
In a study titled "Evaluation of three sets of advanced backcrosses of eggplant with wild relatives from different gene pools under low N fertilization conditions" published in Horticulture Research, 22 morpho-agronomic, physiological, and NUE traits were evaluated under low nitrogen (LN) fertilization conditions in CWRs of eggplant (S. insanum, S. dasyphyllum and S. elaeagnifolium) and their advanced backcrosses (ABs; BC3 to BC5 generations).
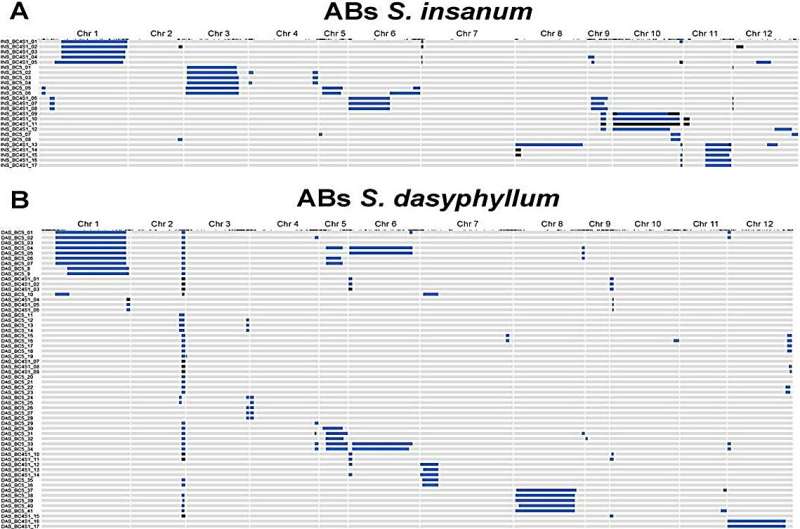
Genome coverage of the donor wild relatives varied, with the highest coverage observed in S. elaeagnifolium at 99.2%. For S. insanum, significant representation was observed on chromosomes 1 (86.8%) and 3 (80.9%), while for S. dasyphyllum, emphasis was on chromosomes 1 (84.8%) and 5 (86.3%).
Upon characterizing S. melongena recurrent parents (MEL5, MEL1, and MEL3), notable disparities emerged between nitrogen treatments. For instance, a 3.7-fold and 5.0-fold change in yield and fruit number (F-Number), respectively, was identified across treatments for MEL5. Additionally, fruit metrics, such as fruit pedicel length in MEL5, exhibited differences under varied nitrogen conditions.
Principal components analysis (PCA) revealed trait groupings among the AB sets, with 48.8% total variation accounted for in the S. insanum and its recurrent parent S. melongena MEL5. Pearson linear correlations showcased significant trait relationships across the AB sets.
A total of 16 putative quantitative trait loci (QTLs) were identified across the AB sets, hinting at underlying genetic controls for specific traits, and potential candidate genes were pinpointed from the eggplant reference genome assembly. Of the 16 putative quantitative trait loci (QTLs) identified, five were localized to the same position on chromosome 9 of S. insanum. The "67/3" eggplant reference genome further pinpointed potential candidate genes, including the nitrate transporter 1/peptide transporter on chromosome 9.
In summary, this research emphasizes the vast potential of eggplant wild relatives for genetic improvement under low nitrogen conditions to promote sustainable agriculture. The identified QTLs and their associations provide a basis for innovative eggplant breeding efforts to support improved yield, quality and nitrogen use efficiency of eggplant under LN conditions.
More information: Gloria Villanueva et al, Evaluation of three sets of advanced backcrosses of eggplant with wild relatives from different gene pools under low N fertilization conditions, Horticulture Research (2023). DOI: 10.1093/hr/uhad141
Journal information: Horticulture Research
Provided by NanJing Agricultural University