This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Elucidating the genetic basis of downy mildew resistance in spinach with skim resequencing: RPF2 and RPF3 loci
The cultivation of spinach (Spinacia oleracea L.), a popular and nutritious vegetable, is majorly threatened by the downy mildew disease. This disease is caused by the host-specific pathogen Peronospora effusa, which has evolved into 19 distinct races.
Selfing of female spinach plants generates inbreed seeds that lack the desired combination of complementing RPF (Resistance to Peronospora farinosa) loci, causing downy mildew infection. As new pathogen races appeared, rendering previously resistant spinach susceptible once again, this has necessitated continuous research into novel RPF genes.
Advanced genetic sequencing has located several RPF loci on the spinach chromosome, aiding in developing resistant strains. The ongoing challenge underscores the need for research to ensure sustainable spinach cultivation.
In April 2023, Horticulture Research published a research paper entitled "Skim resequencing finely maps the downy mildew resistance loci RPF2 and RPF3 in spinach cultivars whale and Lazio".
In this study, the differential cultivars Lazio and Whale (resistant to race 5 of P. effusa) were crossed with Viroflay (susceptible to all races of P. effuse) to generate F1 seeds. The F1 male and female plants were permitted to inter-cross with F2 seeds harvested from female plants and used for inoculation and genetic analyses.
Researchers first employed inoculation experiments to understand the resistance response in spinach progeny to P. effusa race 5, parental lines Lazio, near isogenic line2 (NIL2), and NIL3 displayed resistance, while Viroflay and NIL4 were susceptible. Progeny derived from the Viroflay x Lazio cross exhibited a Mendelian 3:1 segregation ratio for resistance, suggesting regulation by a single dominant gene.
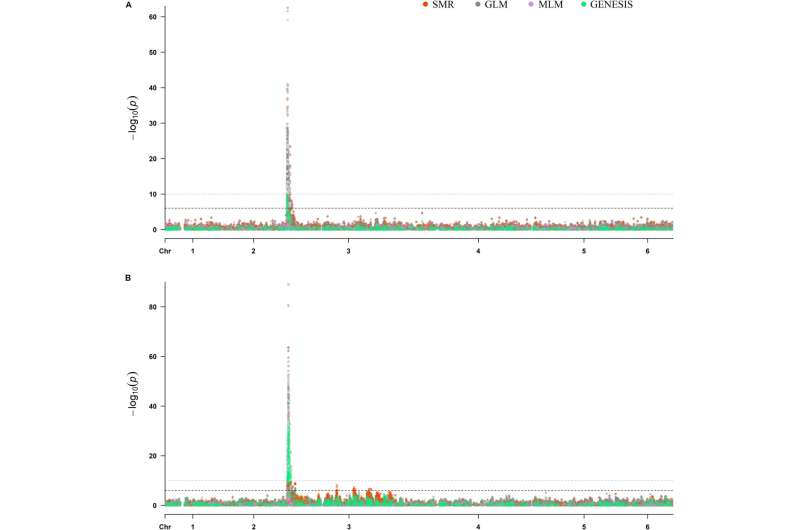
Sequencing of the Viroflay x Lazio progeny population generated 173.79 Gb sequence data, and high-quality sequence reads were aligned against the Sp75 reference genome for single nucleotide polymorphism (SNP) calling. From the initial raw SNP datasets, filtration and imputation processes resulted in 15,021 SNPs across six spinach chromosomes which were deemed suitable for GWAS analysis. Another set of SNPs was also obtained by combining data from the Viroflay x Whale progeny, totaling 34,234 SNPs for downstream GWAS analysis.
In terms of the genetic diversity of the progeny panel, Viroflay x Lazio and Viroflay x Whale differentiated into four subpopulations. These findings were significant for GWAS analysis, aiding in controlling for population structure. The primary goal of this study was to map the RPF2 locus. Through association analysis, 23 SNPs were identified that associated with the RPF2 locus on chromosome 3 between positions 0.47 to 1.46 Mb.
A more extensive study of 384 progeny lines evaluated against P. effusa race 5 pinpointed 28 SNPs in four TASSEL and GENESIS models. The combined GWAS analysis led to the discovery of unique resistance regions for RPF2 in Lazio and RPF3 in Whale. In the quest for associated genes, three significant genes (Spo12793, Spo12908, Spo12916) were identified in close proximity to the RPF2-associated SNPs, plus Spo12908, located at 14–16 Kb away from the peak SNPs (Chr3_1, 221, 009).
In conclusion, this study mapped the RPF2 locus linked to downy mildew resistance within the 0.47 to 1.46 Mb region of chromosome 3 in the Lazio cultivar. The discovery of RPF2-associated genes, especially Spo12821, offers promising prospects for future research to develop downy mildew-resistant spinach cultivars that will benefit spinach producers and consumers alike.
More information: Gehendra Bhattarai et al, Skim resequencing finely maps the downy mildew resistance loci RPF2 and RPF3 in spinach cultivars whale and Lazio, Horticulture Research (2023). DOI: 10.1093/hr/uhad076
Journal information: Horticulture Research
Provided by NanJing Agricultural University