This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Using DNA metabarcoding to analyze soil organism composition change in crop rotation farmland
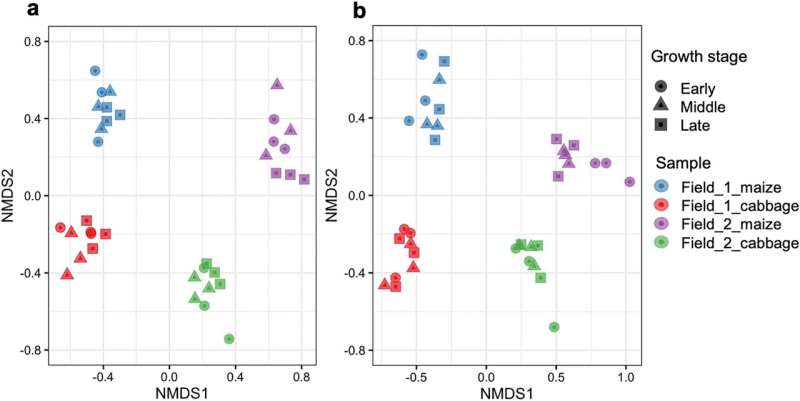
A research team led by Professor Toshihiko Eki of the Department of Applied Chemistry and Life Science, Toyohashi University of Technology and others used DNA metabarcoding to analyze changes in the composition of soil organisms associated with crop growth in two corn-cabbage rotation fields in Tahara City.
The results showed that the soil biota and their networks in agricultural land are unexpectedly easily altered by the soil environment, cultivation history and crops. In the future, the DNA metabarcoding for soil biota analysis will enable the elucidation of the relationship between crops and soil organisms, as well as the evaluation of the biological properties of farmland and prediction of crop diseases, which are expected to contribute to improved crop cultivation technologies.
Historically, soil and humans have been deeply intertwined with each other through agriculture. In recent years, there has been a growing movement internationally to reaffirm the importance of soil and its conservation, as can be seen with the designation of the year 2015 as the International Year of Soil. Diverse organisms such as bacteria, nematodes, arthropods, and fungi exist in soil and form a soil ecosystem, but their actual compositions and dynamics are unclear.
Previously, the research team has analyzed nematodes by fully utilizing the DNA barcoding which identifies biological species based on differences in the organism's unique nucleotide sequences in genes, revealing the unique nematode communities that are adapted to various soil environments. The research team has also introduced the "DNA metabarcoding" using a next-generation sequencer that can decode vast quantities of nucleotide sequences, to analyze all of the pro- and eukaryotes living in soil.
In the present study, the research team applied the DNA metabarcoding to the soil of two fields in Tahara City where corn and cabbage were rotated in 2019 (Field 1 and Field 2) to investigate the taxonomic composition and diversity of pro- and eukaryotes associated with crop growth, interbiotic networks, and the correlation between soil chemical factors and organisms.
The two fields targeted for research had the following differing cultivation history: in the previous year (2018), Field 1 was filled with green manure and not cultivated, whereas Field 2 had the same crop rotation of corn and cabbage. Parts of the 16S and 18S ribosomal RNA genes were used for DNA barcodes to classify pro- and eukaryotes, and a total of 3,086 eukaryotic sequence variants (SVs) and 17,069 prokaryotic SVs were identified as SVs with unique nucleotide sequences.
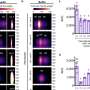
A detailed analysis of the SV data showed that, first, there were biological communities with different compositions in the four soils with different fields and crops, indicating that soil organisms were affected by both fields and crops. Second, the research team investigated the correlation between soil chemical factors and organisms, finding biological groups and major SVs that were closely correlated with soil pH, moisture content, nitrate nitrogen, nutrient salt concentrations, and other factors.
Finally, a network analysis of pro- and eukaryotic SVs showed for the first time that the network structure in the corn-cultivated soil of Field 1, which was not cultivated in the previous year, was clearly less complex than the other soils, but that the network became more complex with subsequent cabbage cultivation. This process largely involved not only fungi but also protists such as Cercozoa.
This research showed that the soil biota and interbiotic networks in agricultural land are influenced and easily changed by the soil environment (e.g., chemical factors), cultivation history, and crops.
This research showed that the DNA metabarcoding approach is useful for the biological analysis of agricultural soils. Recent research has shown that the gut microbiota influences human health, and the relationship between plants (crops) and soil organisms can be compared to the relationship between humans and their gut microbiota.
The research team believes that soil organisms, particularly those living in the rhizosphere, are deeply involved in the health and growth of crops, and they would like to develop their research in the future by focusing on the interactions between plants and rhizosphere organisms.
The research is published in the journal Scientific Reports.
More information: Harutaro Kenmotsu et al, Distinct prokaryotic and eukaryotic communities and networks in two agricultural fields of central Japan with different histories of maize–cabbage rotation, Scientific Reports (2023). DOI: 10.1038/s41598-023-42291-y
Journal information: Scientific Reports
Provided by Toyohashi University of Technology