This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Why S-linked glycosylation cannot adequately mimic the role of natural O-glycosylation

Protein glycosylation is one of the most important post-translational modifications that can be exploited to improve various aspects of therapeutic proteins and industrial enzymes. Different types of glycosylation have a variety of effects on protein properties and functions, and a better understanding of the underlying mechanisms can provide valuable guidance for rational glycoengineering of proteins.
Recently, researchers from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) and their collaborators have revealed why S-glycosylation cannot adequately mimic the role of natural O-glycosylation, and they underscored the pivotal role of the glycosidic linkage in shaping the function of glycosylation.
The results were published in the International Journal of Biological Macromolecules.
In O-glycosylation, the glycan is bound to the oxygen (O) atom of a Ser or Thr side chain. In S-glycosylation, the glycan is bound to the sulfur (S) atom of Cys side chain.
Using S-glycosylation as a replacement for the more commonly occurring O-glycosylation can enhance the resistance of glycans against chemical hydrolysis and enzymatic degradation. These two types of glycosylation exert distinct effects on protein properties and functions.
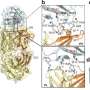
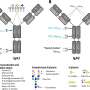
To elucidate the molecular basis behind the observed differences, the researchers conducted a systematic analysis on various glycosylated forms of a model glycoprotein, the carbohydrate-binding module (CBM) of Trichoderma reesei, using nuclear magnetic resonance spectroscopy and molecular dynamic simulations.
Results showed that the S-linked glycosyl exhibited notably greater flexibility compared to the corresponding glycan moiety in O-glycosylation. As a result, glycan-peptide interactions were weakened, leading to significant reduction of stabilizing and substrate binding effects like that of the O-glycosylation.
In addition, they revealed that the altered hydrogen-bonding capability between the glycan and the polypeptide was the main reason for different flexibility between S- and O-glycosylation, which could affect the impact of glycosylation on CBM binding affinity to its substrate by altering the enthalpy and entropy of the binding process. Additionally, they determined the structural and dynamic mechanism of the impact of the second glycosyl on proteins.
"Our study reveals distinct structural and dynamic differences between O- and S-glycosylation, and these differences can lead to significant alterations in the effects of glycosylation," said Prof. Feng Yingang, co-corresponding author of the study. "Caution is imperative when switching glycosylation types in protein glycoengineering."
More information: Chao Chen et al, Structural insight into why S-linked glycosylation cannot adequately mimic the role of natural O-glycosylation, International Journal of Biological Macromolecules (2023). DOI: 10.1016/j.ijbiomac.2023.126649
Provided by Chinese Academy of Sciences