This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
Refining biome labeling for microbial community samples
In a study published in the journal Environmental Science and Ecotechnology, researchers from Huazhong University of Science and Technology have introduced "Meta-Sorter," an AI-based method that leverages neural networks and transfer learning to significantly improve biome labeling for thousands of microbiome samples in the MGnify database, especially those with incomplete information.
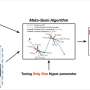
The Meta-Sorter approach comprises two crucial steps. Firstly, a neural network model is meticulously constructed using 118,592 microbial samples from 134 biomes and their respective biome ontology, boasting an impressive average AUROC of 0.896. This model accurately classifies samples with detailed biome information, serving as a strong foundation for further analyses.
Secondly, to address the challenge of newly introduced samples with different characteristics, researchers incorporated transfer learning with 34,209 newly added samples from 35 biomes, including eight novel ones. The transfer neural network model achieved an outstanding average AUROC of 0.989, successfully predicting biome information for newly introduced samples annotated as "Mixed biome."
The results of Meta-Sorter are indeed impressive, achieving an overall accuracy rate of 96.7% in classifying samples among the 16,507 lacking detailed biome annotations. This breakthrough effectively resolves the issue of cascading errors and opens up exciting new possibilities for knowledge discovery across various scientific disciplines, particularly in environmental research.
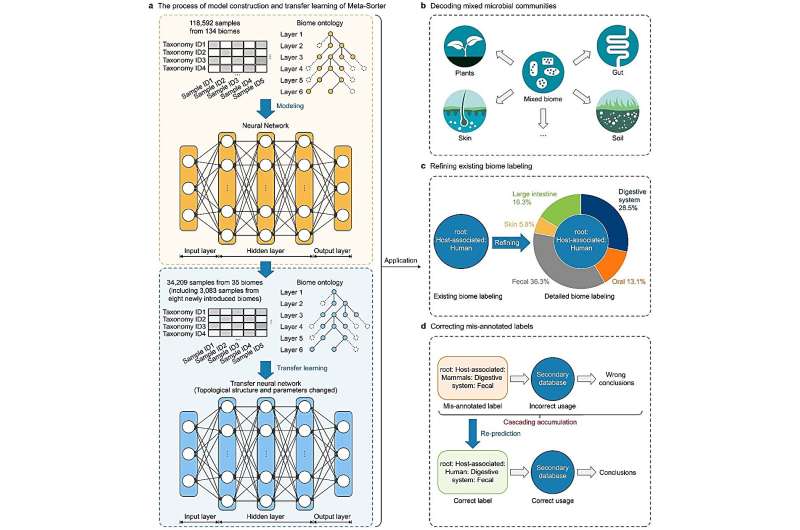
Moreover, Meta-Sorter's success extends to refining the biome annotation for under-annotated and mis-annotated samples. Its intelligent and automatic assignment of precise classifications to ambiguous samples provides valuable insights beyond the original literature, while the differentiation of samples into specific environmental categories enhances the reliability and validity of research conclusions.
With the ongoing development of standardized protocols for data submission and incorporation of additional meta-data information, Meta-Sorter is set to revolutionize the way researchers analyze and interpret microbial community samples. Ultimately, it will lead to more accurate and insightful discoveries in the realm of microbiome research and beyond.
More information: Nan Wang et al, Refining biome labeling for large-scale microbial community samples: Leveraging neural networks and transfer learning, Environmental Science and Ecotechnology (2023). DOI: 10.1016/j.ese.2023.100304
Provided by Chinese Society for Environmental Sciences