This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Behind the rind: New genomic insights into watermelon evolution, quality, and resilience
Watermelon is a globally significant agricultural product, both in terms of the total amount produced and the total economic value generated.
Scientists at the Boyce Thompson Institute have constructed a comprehensive "super-pangenome" for watermelon and its wild relatives, uncovering beneficial genes lost during domestication that could improve disease resistance and fruit quality of this vital fruit crop.
"We aimed to delve deeper into the genetic variations that make watermelons so diverse and unique," stated Professor Zhangjun Fei, the study's lead author. "Our findings not only provide insights into the evolutionary journey of watermelons but also present significant implications for breeding and disease resistance."
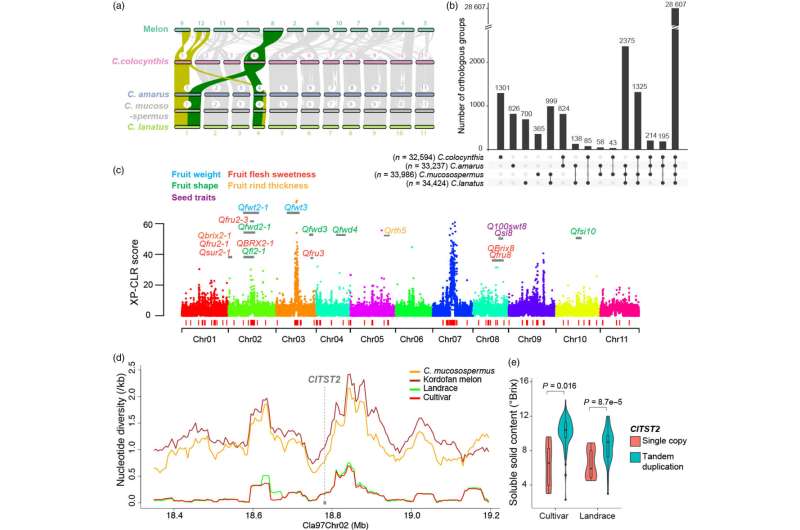
The watermelon super-pangenome was built using reference genome sequences and genome resequencing data from 547 watermelon accessions spanning four species—cultivated watermelon (Citrullus lanatus) and its wild relatives C. mucosospermus, C. amarus, and C. colocynthis.
Analyses of the super-pangenome revealed that many disease-resistance genes present in wild species were lost during domestication, as early farmers selected for fruit quality traits like sweetness, flesh color, and rind thickness. "These beneficial genes could be reintroduced into modern cultivars to breed more resilient watermelon varieties," noted Fei.
A key discovery of the research, recently published in the Plant Biotechnology Journal, was the identification of a tandem duplication of the sugar transporter gene ClTST2 that enhances sugar accumulation and fruit sweetness in cultivated watermelon. This genetic variant was rare in wild watermelons but was selected during domestication.
"The super-pangenome provides a valuable genetic toolkit for breeders and researchers to improve cultivated watermelon," said Fei. "By understanding the genetic makeup and evolutionary patterns of watermelons, we can develop varieties with enhanced yield, increased disease resistance, and improved adaptability."
More information: Shan Wu et al, A Citrullus genus super‐pangenome reveals extensive variations in wild and cultivated watermelons and sheds light on watermelon evolution and domestication, Plant Biotechnology Journal (2023). DOI: 10.1111/pbi.14120
Journal information: Plant Biotechnology Journal
Provided by Boyce Thompson Institute