This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
C. difficile, emerging pathogens, genomics, and antimicrobial resistance
A new study published in OMICS: A Journal of Integrative Biology has identified genes for virulence and antimicrobial resistance in two bacteria that co-occur with C. difficile, suggesting these pathogens as emerging potential threats in planetary health.
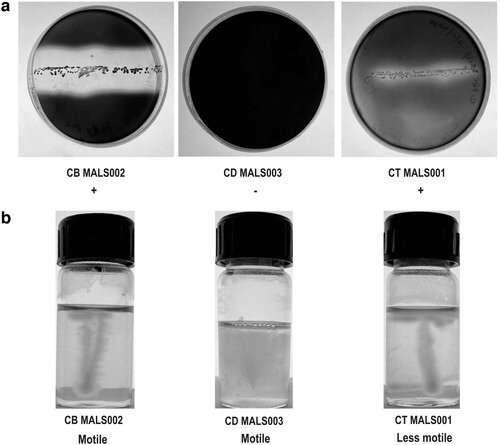
Thokur Sreepathy Murali, Ph.D., Ankit Singh Tanwar, Padival Shruptha and colleagues from Manipal Academy of Higher Education, India, and co-author Angela Brand, MD, Ph.D., MPH from Maastricht University, The Netherlands, performed comparative genome analyses of three Clostridia species, C. difficile, C. butyricum, and C. tertium.
C. difficile can cause diarrhea, colitis, sepsis and multiple organ dysfunction. C. butyricum and C. tertium, which reside in the gut, can harbor toxin/virulence genes, and thus, could pose a threat to human health.
Studies such as this one "will further improve our understanding of development of antimicrobial resistance, provide new avenues in genomic monitoring of emerging pathogens and offer better treatment strategies for crippling infectious diseases," state the investigators.
"The study provides a timely application of comparative genomics in planetary health and opens up new avenues for diagnostics and therapeutics innovation. The findings unpack the potential threats and interactions of three Clostridia species. I welcome new manuscripts on multi-omics applications in planetary health for peer-review in the journal," says Vural Özdemir, MD, Ph.D., DABCP, Editor-in-Chief of OMICS.
More information: Ankit Singh Tanwar et al, Emerging Pathogens in Planetary Health and Lessons from Comparative Genome Analyses of Three Clostridia Species, OMICS: A Journal of Integrative Biology (2023). DOI: 10.1089/omi.2023.0034
Provided by Mary Ann Liebert, Inc