This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A super-resolution microscopy method for rapid differentiation of molecular structures in 3D
Super-resolution microscopy methods are essential for uncovering the structures of cells and the dynamics of molecules. Since researchers overcame the resolution limit of around 250 nanometers (while winning the 2014 Nobel Prize in Chemistry for their efforts), which had long been considered absolute, the methods of microscopy have progressed rapidly.
Now a team led by LMU chemist Prof. Philip Tinnefeld has made a further advance through the combination of various methods, achieving the highest resolution in three-dimensional space and paving the way for a fundamentally new approach for faster imaging of dense molecular structures. The new method permits axial resolution of under 0.3 nanometers.
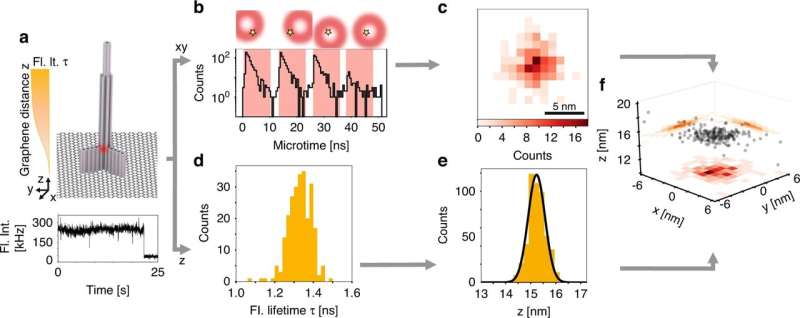
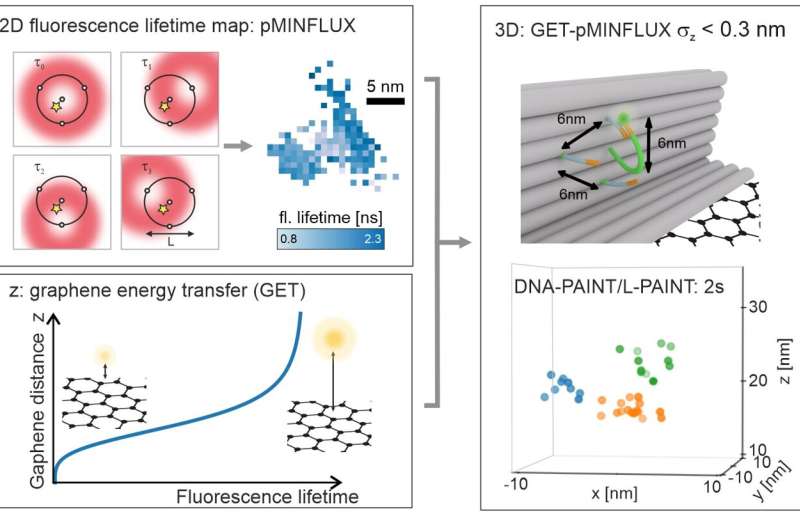
The researchers combined the so-called pMINFLUX method developed by Tinnefeld's team with an approach that utilizes special properties of graphene as an energy acceptor. pMINFLUX is based on the measurement of the fluorescence intensity of molecules excited by laser pulses. The method makes it possible to distinguish their lateral distances with a resolution of just 1 nanometer.
Graphene absorbs the energy of a fluorescent molecule that is no more than 40 nanometers distant from its surface. The fluorescence intensity of the molecule therefore depends on its distance from graphene and can be used for axial distance measurement.
DNA-PAINT increases the speed
Consequently, the combination of pMINFLUX with this so-called graphene energy transfer (GET) furnishes information about molecular distances in all three dimensions—and does this in the highest resolution achievable to date of under 0.3 nanometers. "The high precision of GET-pMINFLUX opens the door to new approaches for improving super-resolution microscopy," says Jonas Zähringer, lead author of the paper.
The researchers also used this to further increase the speed of super-resolution microscopy. To this end, they drew on DNA nanotechnology to develop the so-called L-PAINT approach. In contrast to DNA-PAINT, a technique that enables super-resolution through the binding and unbinding of a DNA strand labeled with a fluorescent dye, the DNA strand in L-PAINT has two binding sequences.
In addition, the researchers designed a binding hierarchy, such that the L-PAINT DNA strand binds longer on one side. This allows the other end of the strand to locally scan the molecule positions at a rapid rate.
"As well as increasing the speed, this permits the scanning of dense clusters faster than the distortions arising from thermal drift," says Tinnefeld. "Our combination of GET-pMINFLUX and L-PAINT enables us to investigate structures and dynamics at the molecular level that are fundamental to our understanding of biomolecular reactions in cells."
The findings are published in the journal Light: Science & Applications.
More information: Jonas Zähringer et al, Combining pMINFLUX, graphene energy transfer and DNA-PAINT for nanometer precise 3D super-resolution microscopy, Light: Science & Applications (2023). DOI: 10.1038/s41377-023-01111-8
Journal information: Light: Science & Applications
Provided by Ludwig Maximilian University of Munich