This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Hijacking our cells' enzymes to eliminate disease-causing proteins
By studying how enzymes move from one membrane compartment to another inside a cell, scientists at the University of Illinois Chicago have figured out a way to better target cellular proteins, which play a role in many diseases.
Their findings, published in a Cell Reports paper titled "Palmitoylation and PDE6δ regulate membrane-compartment-specific substrate ubiquitylation and degradation," have implications for developing new therapies.
Lead author Shafi Kuchay, assistant professor of biochemistry and molecular genetics in the College of Medicine and member of the University of Illinois Cancer Center at UIC, said that most common drugs work by targeting proteins that are located at the membranes of cells. Many of these proteins can cause diseases by being overly active. Unfortunately, most currently available drugs just block the activity of the harmful proteins, and while they are helpful in the short term, resistance to the drugs can develop over time.
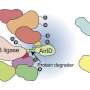
"We are interested in understanding how and why ubiquitin ligase enzymes, which can naturally degrade proteins, move around the cellular compartments and are able to find very specific proteins to degrade," Kuchay said. "We want to leverage this natural process so we can repurpose ubiquitin ligase enzymes to completely remove problematic proteins that lead to diseases, as opposed to just blocking their activity."
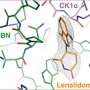
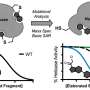
The researchers looked at a ubiquitin ligase enzyme named FBXL2, known to degrade proteins at various cellular membrane compartments. They found that by attaching or detaching a fat molecule or lipid to FBXL2—a process called palmitoylation and de-palmitoylation—they could direct where the FBXL2 went.
Using animal cells, they found that FBXL2 would move to degrade proteins in a particular cell membrane compartment if the lipid was attached. In the detached state, FBXL2 would move to other compartments. They also discovered that in order to travel in the aqueous cellular environment for the delivery of lipid-modified FBXL2 to membrane compartments, it used a trafficking protein called PDE6D, which is known to shield the lipid modifications.
"We can engineer this lipid modification signal to direct the ubiquitin ligase to induce protein degradation in a specific compartment of the cell, which we predict has potential to make drugs safer and more effective," Kuchay said.
Together, these findings mean that drug actions can be restricted to specific regions of a cell and ubiquitin ligase enzymes can be better directed to problematic proteins to destroy them for improving drug efficacy. The researchers think that hijacking these ligases has particular relevance in designing treatments for certain types of cancer or neurodegenerative diseases.
UIC student David Liang, an undergraduate in the College of Liberal Arts and Sciences, is the first author of the paper. Additional co-authors include Liping Jiang, Sameer Ahmed Bhat, Sonia Missiroli, Mariasole Perrone, Angela Lauriola, Ritika Adhikari, Anish Gudur, Zahra Vasi, Ian Ahearn, Daniele Guardavaccaro, Carlotta Giorgi and Mark Philips.
More information: David Liang et al, Palmitoylation and PDE6δ regulate membrane-compartment-specific substrate ubiquitylation and degradation, Cell Reports (2023). DOI: 10.1016/j.celrep.2023.111999
Journal information: Cell Reports
Provided by University of Illinois at Chicago