This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
A transnational collaboration leads to the characterization of an emergent plant virus
In the 21st century, "collaboration" has become a popular buzzword, but effectively working together across disciplines and countries is easier said than done. However, authentic collaboration is critical to the fight against plant pathogens; sharing information on plant diseases facilitates early detection, efficient and rapid characterization, and subsequent management.
Physostegia chlorotic mottle virus (PhCMoV), a plant disease first identified in Austria in 2018, initially received inadequate characterization. This then sparked studies across Europe as new symptoms emerged on economically important crops. These independent studies converged into one study, published in Plant Disease, demonstrating the power of collaboration beyond a mere buzzword.
In the coalescent study, Coline Temple and colleagues from eight laboratories across five European countries utilized prepublication sharing of high throughput sequencing (HTS) data to improve knowledge on PhCMoV biology, epidemiology, and genetic diversity.
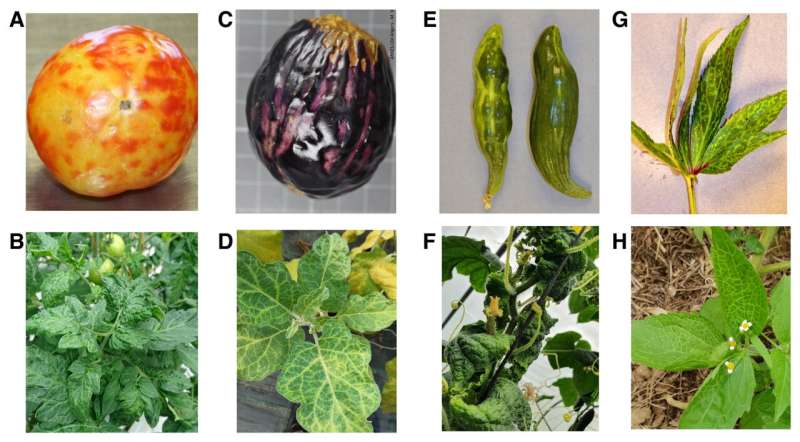
The researchers identified PhCMoV in eight European countries, in addition to Austria, and confirmed its presence in samples collected in 2002. Mechanically inoculating PhCMoV to healthy host plants in control conditions enabled the authors to validate the association of the virus with symptoms. Their results show that PhCMoV can infect at least nine plant species and cause severe fruit symptoms on economically important crops such as tomato, eggplant, and cucumber.
Additionally, sequencing 21 different infected host plants from various origins allowed for comparison of genomes, which showed that the genomes of two isolates from the same site barely evolved over 17 years. This suggests that the same isolate of the microorganism can survive without adaptation in a specific ecosystem.
In addition to characterizing an emergent plant disease, this study illustrates how solidarity and trust between scientists can accelerate a common goal. Corresponding Author Sébastien Massart recalls the process, commenting, "The most exciting moment of this research was when we progressively realized that we were not alone, that different research groups had suddenly and independently detected this virus in different symptomatic host plants and countries all over Europe."
Uniting against this virus by sharing HTS data prepublication created a collaborative space free from competition, which shows how "trusting colleagues and sharing information on the most recent results between groups can facilitate synergy, accelerate virus characterization, and gather the most information in a single publication," Massart says.
This research will ultimately benefit all plant health stakeholders and offer a solid foundation of knowledge for further studies on plant rhabdoviruses.
More information: Coline Temple et al, Biological and Genetic Characterization of Physostegia Chlorotic Mottle Virus in Europe Based on Host Range, Location, and Time, Plant Disease (2022). DOI: 10.1094/PDIS-12-21-2800-RE
Provided by American Phytopathological Society