Coronavirus drug target that could halt virus replication identified
Structural details of an attractive drug target in coronaviruses that could be used against SARS-CoV-2 and in future pandemics have been published by international teams co-led by UCL researchers.
Two new studies published in the International Journal of Molecular Sciences and eLife reveal pockets in an important piece of the virus' machinery that drugs could bind to in order to halt virus replication.
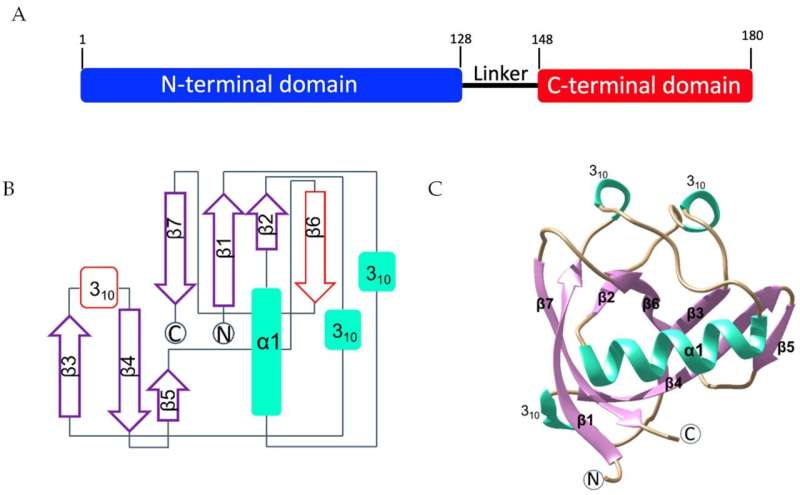
One of the proteins known to play a role in infection with SARS-CoV-2 (the virus responsible for the disease, COVID-19) is non-structural protein-1 (Nsp1). Nsp1 is found in several coronaviruses such as SARS-CoV-2, MERS and SARS, and its role is to help the virus hijack the human body's protein production machinery.
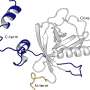
In the International Journal of Molecular Sciences study, an international team led by Professor Frank Kozielski (UCL School of Pharmacy) used state-of-the-art technology to identify ligands (a type of binding molecule) binding to SARS-CoV-2 Nsp1. To do this they grew hundreds of protein crystals which were then exposed to chemical compounds.
The team identified and characterized two novel ligand binding sites on SARS-CoV-2 Nsp1. They also showed that differences in these sites exist between the three medically relevant coronaviruses infecting humans.
First author Shumeng Ma, a Ph.D. student at the UCL School of Pharmacy, said, "This study was an example of how various scientists from different disciplines came together to work on the common goal to contribute to the understanding of Nsp1 and its characterization as a potential drug target."
In the eLife study, a team led by University of Geneva and UCL Chemistry scientists explored whether it might be possible to design drugs against Nsp1 using computational methods. The team used computational models to study its 3D structure and how it changes shape under different conditions or when attached to a variety of molecules. This revealed four previously unidentified binding pockets, two of which were fully hidden, and two that were partially hidden.
First author Alberto Borsatto, a Ph.D. student at the University of Geneva, said, "Nsp1 is an attractive antiviral drug target in principle, but the shape of Nsp1 makes designing a potential drug difficult. So far, only shallow, superficial cavities have been seen on the Nsp1 surface, and this makes it challenging for drugs to attach and interfere with Nsp1's function."
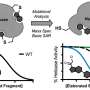
To determine whether these pockets could be targeted with drugs, the team conducted an experiment where they used computational methods to screen a library of 1,000 fragments and identified 59 different chemical fragments that had 'attached' to the protein in the computer model. To their surprise, only one of the fragments experimentally bound to Nsp1.
To see whether these observations would only be true for SARS-CoV-2, the team looked at the structures of Nsp1 proteins from other coronaviruses. Their computer models suggest that ligands targeting any of the pockets in SARS-Cov-2 Nsp1 could also target the corresponding pockets in other coronaviruses tested. This offers the potential to develop drugs that could protect against future coronavirus pandemics.
Co-senior author of the eLife paper, Professor Francesco Luigi Gervasio (UCL Chemistry and University of Geneva), said, "We've characterized potential drug binding pockets in the SARS-CoV-2 virus Nsp1 and predicted four partially hidden pockets, one of which we've validated using X-ray crystallography. The results of this research can be used as a stepping stone for the design of Nsp1 inhibitors for SARS-CoV-2 and, potentially, for other coronaviruses."
More information: Shumeng Ma et al, Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening, International Journal of Molecular Sciences (2022). DOI: 10.3390/ijms232012448
Alberto Borsatto et al, Revealing druggable cryptic pockets in the Nsp1 of SARS-CoV-2 and other β-coronaviruses by simulations and crystallography, eLife (2022). DOI: 10.7554/eLife.81167
Journal information: eLife
Provided by University College London