Researchers investigate neuron differentiation in fruit fly brains
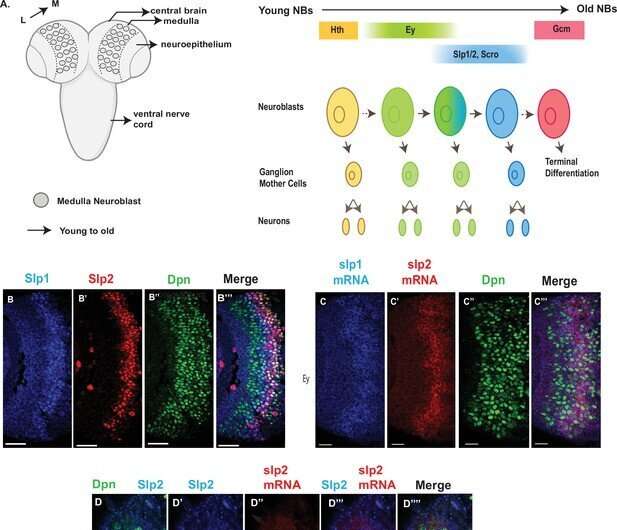
The brains of all higher-order animals are filled with a diverse array of neuron types, with specific shapes and functions. Yet, when these brains form during embryonic development, there is initially only a small pool of cell types to work with. So how do neurons diversify over the embryo's development? Researchers know that neural stem cells called neuroblasts divide multiple times to sequentially produce neurons of specialized function, but the mechanisms of this process and how the timing varies for different genes and neuron types are still not fully understood.
In a new paper published in eLife, Alokananda Ray, a Ph.D candidate during the time of the study and now graduated, and Xin Li (GNDP), an assistant professor of cell and developmental biology at the University of Illinois Urbana-Champaign, shed light on the process in the optic medulla of Drosophila melanogaster, the fruit fly.
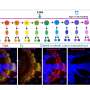
As neuroblasts divide and differentiate, they express transcription factors which ultimately direct the daughter cells on what kind of neuron to be. Because they are expressed in a particular way depending on when they split, these transcription factors, called temporal transcription factors, act as a marker that tells researchers at which specific stage the neuroblast is, and allows them to piece together the order of events in this neurogenesis cascade. The researchers focused on two different TTFs in the fruit fly brain, called eyeless and sloppy-paired, to better understand how differences in the expression of TTFs that lead to different neuron fates.
"Nervous systems diversify from a small pool of neural stem cells to the great diversity of neurons we see in adult brains of higher-ordered animals," said Ray. "We really wanted to understand the molecular mechanisms that drive the transition of these neuroblasts from expressing one temporal transcription factor to the next transcription factor, which ultimately determines what type of neurons these progenies will become."
The researchers used genetics and a number of techniques including reporter assays, antibody staining and microscopy to measure the expression pattern of genes within the optic medulla of fruit fly brains during development. Typically, the regions of the DNA that are considered to be "important" are the sequences that contain genes. However, through these experiments, the researchers discovered that two non-coding regions near the sloppy-paired genes were essential to making sure the sloppy-paired TTFs expressed at the right time and amount. Researchers then removed these non-coding DNA regions, called enhancers, using the gene-editing technique CRISPR to see how the brain of the flies were affected, and found that flies with deleted enhancers showed a complete absence of expression of the sloppy-paired TTF in medulla neuroblasts.
"On the outside, we don't see morphological changes from removing sloppy-paired enhancers, but neurons generated in the sloppy-paired stage will be missing from the brain, and I think the neurons generated in later stages will also be lost," said Li.
The second major finding in the paper was that a mechanism called Notch signaling works together with the preceding TTFs to activate the expression of the next TTFs in question. The researchers determined that not only is Notch signaling important for regulating TTF expression, but the way it regulates is dependent on where in the neurogenesis cascade the cells are. In other words, once a certain number of a specific neuron type have been made, Notch signaling regulates the transition such that the neuroblasts begin differentiating into a different neuron type.
"One TTF is required to activate the next TTF, but that alone is not sufficient to cause the transition," explained Li. "After each cell cycle, Notch signaling will further activate the next TTF until a certain level is reached, at which point it will repress the previous TTF; then the transition to the next TTF stage will happen. Basically, this mechanism couples the temporal patterning in these neural stem cells with the generation of the appropriate number of neurons at each temporal stage."
Though TTFs vary between animals, Notch signaling is highly conserved, meaning that understanding the molecular mechanisms that regulate neuron differentiation in the fly can potentially translate across other higher-order animals. The findings in this study illuminate some of the mechanisms underlying neuron diversity in the brain, but the researchers said there is more to be explored.
"Identifying the molecular determinants, or enhancers, that are required for the transition to take place from eyeless to sloppy-paired gives us ideas for how other transitions may also be regulated," Ray explained. "We're going to try to identify other enhancers that previous TTFs bind to activate the expression of subsequent factors."
More information: Alokananda Ray et al, A Notch-dependent transcriptional mechanism controls expression of temporal patterning factors in Drosophila medulla, eLife (2022). DOI: 10.7554/eLife.75879
Journal information: eLife
Provided by University of Illinois at Urbana-Champaign