Molecular fingerprint behind beautiful pearls revealed

Pearl oysters are an important aquaculture animal in Japan, as they produce the beautiful pearls that are sought after for necklaces, earrings, and rings. In the early 1990s, this aquaculture industry was bringing in around 88bn yen annually. But, in the last 20 years, a combination of new diseases and red tides has seen production of Japan's pearls drop from around 70,000 kg a year to just 20,000 kg.
Now, researchers from the Okinawa Institute of Science and Technology (OIST), in collaboration with a number of other research institutes including K. MIKIMOTO & CO., LTD, Pearl Research Institute and Japan Fisheries Research and Education Agency, have constructed a high-quality, chromosome-scale genome of pearl oysters, which they hope can be used to find resilient strains. The research was published recently in DNA Research.
"It's very important to establish the genome," said one of the two first authors, Dr. Takeshi Takeuchi, staff scientist in OIST's Marine Genomics Unit. "Genomes are the full set of an organism's genes—many of which are essential for survival. With the complete gene sequence, we can do many experiments and answer questions around immunity and how the pearls form."
In 2012, Dr. Takeuchi and his collaborators published a draft genome of the Japanese pearl oyster, Pinctada fucata, which was one of the first genomes assembled of a mollusk. They continued genome sequencing in order to establish a higher quality, chromosome-scale genome assembly.
Dr. Takeuchi went on to explain that the oyster's genome is made up of 14 pairs of chromosomes, one set inherited from each parent. The two chromosomes of each pair carry nearly identical genes, but there can be subtle differences if a diverse gene repertoire benefits their survival.
Traditionally, when a genome is sequenced, the researchers merge the pair of chromosomes together. This works well for laboratory animals, which normally have almost identical genetic information between the pair of chromosomes. But for wild animals, where a considerable number of variants in genes exist between chromosome pairs, this method leads to a loss of information.
In this study, the researchers decided not to merge the chromosomes when sequencing the genomes. Instead, they sequenced both sets of chromosomes—a method that is very uncommon. In fact, it's probably the first research focused on marine invertebrates to use this method.
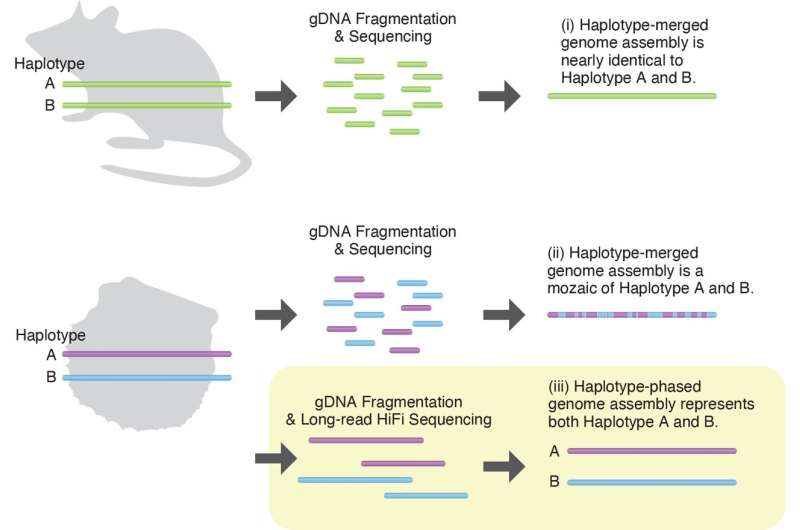
As pearl oysters have 14 pairs of chromosomes, they have 28 in total. OIST researchers Mr. Manabu Fujie and Ms. Mayumi Kawamitsu used state-of-the-art technology to sequence the genome. The other first author, Dr. Yoshihiko Suzuki, former Postdoctoral Scholar in OIST's Algorithms for Ecological and Evolutionary Genomics and now at the University of Tokyo, and Dr. Takeuchi reconstructed all 28 chromosomes and found key differences between the two chromosomes of one pair—chromosome pair 9. Notably, many of these genes were related to immunity.
"Different genes on a pair of chromosomes is a significant find because the proteins can recognize different types of infectious diseases," said Dr. Takeuchi.
He pointed out that when the animal is cultured, there is often a strain that has a higher rate of survival or produces more beautiful pearls. The farmers often breed two animals with this strain but that leads to inbreeding and reduces genetic diversity. The researchers found that after three consecutive inbreeding cycles, the genetic diversity was significantly reduced. If this reduced diversity occurs in the chromosome regions with genes related to immunity, it can impact the immunity of the animal.
"It is important to maintain the genome diversity in aquaculture populations," concluded Dr. Takeuchi.
More information: A high-quality, haplotype-phased genome reconstruction reveals unexpected haplotype diversity in a pearl oyster, DNA Research (2022). DOI: 10.1093/dnares/dsac035
Provided by Okinawa Institute of Science and Technology