Mathematical model could bring us closer to effective stem cell therapies
Stem cells are the very definition of potential: they have contained in their DNA the potential to become virtually any cell in the body. Scientists have been working for decades to harness this power to use as medicine—think replacing damaged cells with brand new ones—that could treat or even cure everything from diabetes to heart disease.
However, so-called regenerative medicine is still in its early days and very few stem cell therapies have been found to be effective. Part of the problem is scientists still don't fully understand how a stem cell transforms into its final form, be it a blood cell or heart cell. Without a clear understanding of that process, scientists can't control it to use as a therapy.
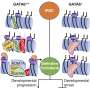
One of the lingering questions is how a cell's genes are expressed over time, from DNA to RNA to protein. The genome doesn't simply follow the instructions encoded in its DNA; it responds to signals from the environment that tell it what to express and when to express it, by making alterations to the structure of chromatin, the tightly wound bundle that contains the DNA. These chemical signals are called the epigenome.
Furthermore, for a gene to be expressed, its DNA must be transcribed into RNA. The readout of a cell's RNA is called the transcriptome.
"The big question in the field is which changes first, the epigenome or the transcriptome," said Joshua Welch, Ph.D., an assistant professor in the Department of Computational Medicine and Bioinformatics at the U-M Medical School. A study from his team, published in Nature Biotechnology, provides a mathematical model that may be used to estimate this timing.
Until recently, researchers could not see gene expression in an individual cell. Thanks to single cell sequencing techniques, they now can. But the timing of changes is still hard to visualize, as measuring the cell destroys it.
"To address this, we developed an approach based on models in basic physics," explained Welch, "treating the cells like they are masses moving through space and we are trying to estimate their velocity."
The model, dubbed MultiVelo, predicts the direction and speed of the molecular changes the cells are undergoing.
"Our model can tell us which things are changing first—epigenome or gene expression—and how long it takes for the first to ramp up the second," said Welch.
They were able to verify the method using four types of stem cells from the brain, blood and skin, and identified two ways in which the epigenome and transcriptome can be out of sync. The technique provides an additional, critical layer of insight to so called cellular atlases, which are being developed using single cell sequencing to visualize the various cell types and gene expression in different body systems.
By understanding the timing, Welch noted, researchers are closer to steering the development of stem cells for use as therapeutics.
"One of the big problems in the field is the artificially differentiated cells created in the lab never quite make it to full replicas of their real-life counterparts," said Welch. "I think the biggest potential for this model is better understanding what are the epigenetic barriers to fully converting the cells into whatever target you want them to be."
Additional authors on this paper include Chen Li, Maria C. Virgilio, and Kathleen L. Collins.
More information: Chen Li et al, Multi-omic single-cell velocity models epigenome–transcriptome interactions and improves cell fate prediction, Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01476-y
Journal information: Nature Biotechnology
Provided by University of Michigan