Discovering new cancer treatments in the 'dark matter' of the human genome
Cancer is the second leading cause of death in Switzerland. Among the different types of cancers, non-small cell lung cancer (NSCLC) kills the most patients and remains largely incurable. Unfortunately, even newly approved therapies can extend the life of patients by only a few months and only few survive the metastatic stadium long-term. Thus, new treatments that attack the cancer in novel ways are sought.
In a recently published study in the journal Cell Genomics, researchers of the University of Bern and the Insel Hospital have determined new targets for drug development for this cancer type.
The dark matter of the genome
For new targets, they looked at the poorly-understood class of genes called "long noncoding RNAs (ribonucleic acids)" (lncRNAs). LncRNAs exist in abundance in the so-called "dark matter" or non-protein-coding DNA that constitutes the vast majority of our genome. The human genome contains around 20,000 "classical" protein-coding genes, but this number is dwarfed by 100,000 lncRNAs. The biological functions of 99% of lncRNAs are unknown.
As their name "long noncoding RNAs" implies, unlike messenger RNAs (mRNAs), lncRNAs do not encode the construction plans for proteins. As in mRNAs, the building instructions for lncRNAs are contained in the cell's DNA.
New tool determines potential targets
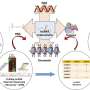
To study the role of lncRNAs in NSCLC, the researchers started by analyzing publicly available datasets to see which lncRNAs are present in NSCLC. This analysis led to a list of over 800 lnRNAs, whose importance for NSCLC cells the researchers wished to investigate. For this investigation, they developed a screening system that prevents the production of the selected lncRNAs by deleting part of their construction instructions in the DNA.
The researchers applied their screening system to two NSCLC cell lines derived from patients, and looked at how the inhibition of the selected lncRNAs affected so called "hallmarks" of cancer cells. Hallmarks are cellular behaviors that contribute to disease progression: proliferation, metastasis formation and therapy resistance.
"The advantage of assessing three different cancer hallmarks is that we have a comprehensive view but also have substantial amounts of data from different experiments, from which we needed to derive a single list of long noncoding RNAS that are important for non-small cell lung cancer," says Rory Johnson, Assistant Professor at the University of Bern, who led the NCCR RNA & Disease project. The analysis yielded, in the end, a list of 80 high-confidence candidate lncRNAs important for NSCLC, out of the over 800 investigated. From these 80, the researchers picked out several lncRNAs for follow-up experiments.
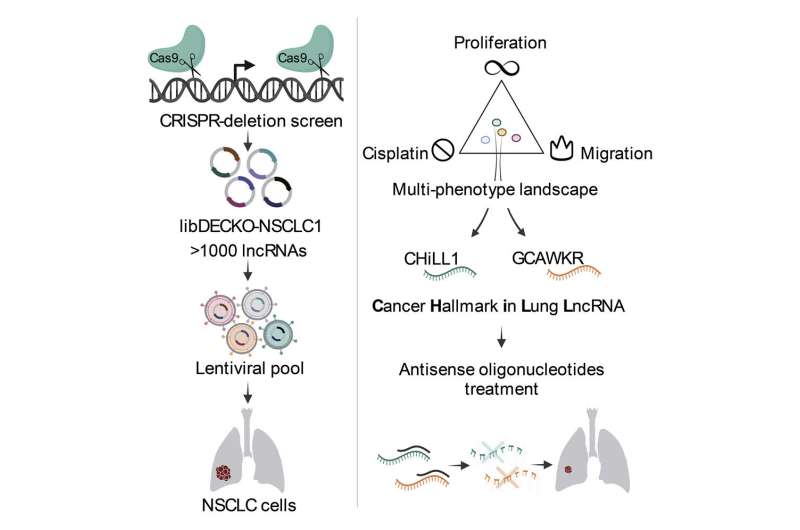
Destroying a long RNA with a short one
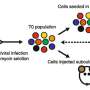
For these follow-up experiments the researchers used an approach that does not work at the DNA level, but targets lncRNAs after their production. For this purpose, the researchers used small chemically-synthesized RNAs called antisense oligonucleotides (ASOs), which bind to the lncRNAs they target and lead to their degradation. Of note, several ASOs are approved for treating human diseases, although none yet for cancer.
These follow up experiments showed that for the majority of the picked lncRNAs, their destruction by an ASO inhibited cancer cell division in cell culture. Importantly, the same treatment produced little if any effect on non-cancerous lung cells, which should not be harmed by the cancer treatment. In a 3D model of NSCLC, which more closely resembled the tumor than cell culture, the inhibition of a single lncRNA with an ASO reduced the tumor growth by more than half.
"We were positively surprised to see how well the antisense oligonucleotides could restrain tumor growth in different models," states Taisia Polidori, co-first author, who worked on the project as part of her doctoral thesis research at the University of Bern.
Therapy development and application to other tumor types
The researchers are continuing their research in pre-clinical cancer models and are considering collaborating with existing companies or creating a startup in order to develop a drug to treat patients.
Regarding other cancers, Roberta Esposito, co-first author and postdoc at the University of Bern, said, "Like a telescope that can be quite easily repositioned to study a different part of space, our approach should be easily adaptable to reveal new potential treatment types for other cancer types." Dr. Esposito will now apply the "telescope" to identify new targets for colorectal cancer.
More information: Roberta Esposito et al, Multi-hallmark long noncoding RNA maps reveal non-small cell lung cancer vulnerabilities, Cell Genomics (2022). DOI: 10.1016/j.xgen.2022.100171
Provided by University of Bern