Using math to predict SARS-CoV-2 protein mutations
Oak Ridge National Laboratory, the University of Tennessee at Chattanooga and Tuskegee University researchers used mathematics to predict which areas of the SARS-CoV-2 spike protein are most likely to mutate.
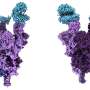
In a study published in Polymers, the team analyzed the topology of more than 13,000 Protein Data Bank structures and found that mutations in the virus's spike protein were most likely to occur in areas with high topological free energy, which makes a protein less stable.
The SARS-CoV-2 omicron variant mutated in an area consistent with researchers' calculations, affirming the paper's results.
This methodology allows scientists to predict where protein mutations may occur using mathematics and could be applied to any protein, potentially opening the door for better understanding of viral dynamics and more efficient drug therapies.
"We believe the approach is virus independent," said ORNL's Bobby Sumpter. "It's all based upon the topology of proteins."
More information: Quenisha Baldwin et al, The Local Topological Free Energy of the SARS-CoV-2 Spike Protein, Polymers (2022). DOI: 10.3390/polym14153014
Provided by Oak Ridge National Laboratory