An AI message decoder based on bacterial growth patterns

From a box of Cracker Jack to The Da Vinci Code, everybody enjoys deciphering secret messages. But biomedical engineers at Duke University have taken the decoder ring to place it's never been before—the patterns created by bacterial colonies.
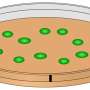
Depending on the initial conditions used, such as nutrient levels and space constraints, bacteria tend to grow in specific ways. The researchers created a virtual bacterial colony and then controlled growth conditions and the numbers and sizes of simulated bacterial dots to create an entire alphabet based on how the colonies would look after they fill a virtual Petri dish. They call this encoding scheme emorfi.
The encoding is not one-to-one, as the final simulated pattern corresponding to each letter is not exactly the same every time. However, the researchers discovered that a machine learning program could learn to distinguish between them to recognize the letter intended.
"A friend may see many images of me over the course of time, but none of them will be exactly the same," explained Lingchong You, professor of biomedical engineering at Duke. "But if the images are all consistently reinforcing what I generally look like, the friend will be able to recognize me even if they're shown a picture of me they've never seen before."
To encrypt real messages, the encoder ends up creating a movie of a series of patterns, each correlating to a different letter. While they may look similar to the untrained eye, the computer algorithm can distinguish between them. So long as the receiver knows the set of initial conditions that led to their creation, an interloper should not be able to crack the code without a powerful AI of their own.
Give the cypher a try yourself. You can type in anything from your name to the Gettysburg Address, or even the Christmas classic, "Be sure to drink your Ovaltine."
More information: Jia Lu et al, Distributed information encoding and decoding using self-organized spatial patterns, Patterns (2022). DOI: 10.1016/j.patter.2022.100590
Journal information: Patterns
Provided by Duke University