Research team first to develop 3D structure of twinkle protein
Researchers from the National Institutes of Health have developed a three-dimensional structure that allows them to see how and where disease mutations on the twinkle protein can lead to mitochondrial diseases. The protein is involved in helping cells use energy our bodies convert from food. Prior to the development of this 3D structure, researchers only had models and were unable to determine how these mutations contribute to disease. Mitochondrial diseases are a group of inherited conditions that affect 1 in 5,000 people and have very few treatments.
"For the first time, we can map the mutations that are causing a number of these devastating diseases," said lead author Amanda A. Riccio, Ph.D., and researcher in the National Institute of Environmental Health Sciences (NIEHS) Mitochondrial DNA Replication Group, which is part of NIH. "Clinicians can now see where these mutations lie and can use this information to help pinpoint causes and help families make choices, including decisions about having more children."
The new findings will be particularly relevant for developing targeted treatments for patients who suffer from mitochondrial diseases such as progressive external ophthalmoplegia, a condition that can lead to loss of muscle functions involved in eye and eyelid movement; Perrault syndrome, a rare genetic disorder that can cause hearing loss; infantile-onset spinocerebellar ataxia, a hereditary neurological disorder; and hepatocerebral mitochondrial DNA (mtDNA) depletion syndrome, a hereditary disease that can lead to liver failure and neurological complications during infancy.
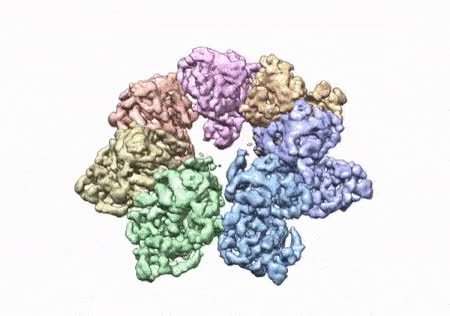
The paper that appears in the Proceedings of the National Academy of Sciences showcases how the NIEHS researchers were the first to accurately map clinically relevant variants in the twinkle helicase, the enzyme that unwinds the mitochondrial DNA double helix. The twinkle structure and all the coordinates are now available in the open data Protein Data Bank that is freely available to all researchers.
"The structure of twinkle has eluded researchers for many years. It's a very difficult protein to work with," noted William C. Copeland, Ph.D., who leads the Mitochondrial DNA Replication Group and is the corresponding author on the paper. "By stabilizing the protein and using the best equipment in the world we were able to build the last missing piece for the human mitochondrial DNA replisome."
The researchers used cryo-electron microscopy (CryoEM), which allowed them to see inside the protein and the intricate structures of hundreds of amino acids or residues and how they interact.
Mitochondria, which are responsible for energy production, are especially vulnerable to mutations. mtDNA mutations can disrupt their ability to generate energy efficiently for the cell. Unlike other specialized structures in cells, mitochondria have their own DNA. In a cell's nucleus there are two copies of each chromosome, however in the mitochondria there could be thousands of copies of mtDNA. Having a high number of mitochondrial chromosomes allows the cell to tolerate a few mutations, but accumulation of too many mutated copies leads to mitochondrial disease.
To conduct the study, the researchers used a clinical mutation, W315L, known to cause progressive external ophthalmoplegia, to solve the structure. Using CryoEM, they were able to observe thousands of protein particles appearing in different orientations. The final structure shows a multi-protein circular arrangement. They also used mass spectrometry to verify the structure and then did computer simulations to understand why the mutation results in disease.
Within twinkle, they were able to map up to 25 disease-causing mutations. They found that many of these disease mutations map right at the junction of two protein subunits, suggesting that mutations in this region would weaken how the subunits interact and make the helicase unable to function.
"The arrangement of twinkle is a lot like a puzzle. A clinical mutation can change the shape of the twinkle pieces, and they may no longer fit together properly to carry out the intended function," Riccio explained.
"What is so beautiful about Dr. Riccio and the team's work is that the structure allows you to see so many of these disease mutations assembled in one place," said Matthew J. Longley, Ph.D., an author and NIEHS researcher. "It is very unusual to see one paper that explains so many clinical mutations. Thanks to this work, we are one step closer to having information that can be used to develop treatments for these debilitating diseases."
More information: Amanda A. Riccio et al, Structural insight and characterization of human Twinkle helicase in mitochondrial disease, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2207459119
Journal information: Proceedings of the National Academy of Sciences
Provided by National Institutes of Health