New study reveals computation-guided approach to suppressing cancer tumor growth
A new study, led by researchers from the University of California, Irvine and the University of California, San Diego, reveals a new computation-guided approach to identify small molecules that can restore aspects of wild-type p53 tumor suppression function to mutated p53, which play an important role in many human cancers. This approach was successful both in vitro and in vivo. This strategy can increase chemical diversity of p53 corrector molecules for clinical development.
The tumor suppressor p53 is one of the most powerful mechanisms organisms use to protect themselves from cancer. Elephants have multiple copies of the p53 gene and rarely get cancer. Humans have only one copy and it is the most mutated gene found in human cancer. Diverse therapeutic approaches are actively pursued to target this pathway.
"Interestingly, a large fraction of p53 alterations are missense mutations, where the genetic code of the p53 is altered in a way that produces a different amino acid than it would normally," explained Peter Kaiser, Ph.D., professor and chair of the Department of Biological Chemistry at the UCI School of Medicine. "This results in abundance of mutant p53 protein levels in tumors that are, in principle, amenable to a corrector drug approach."
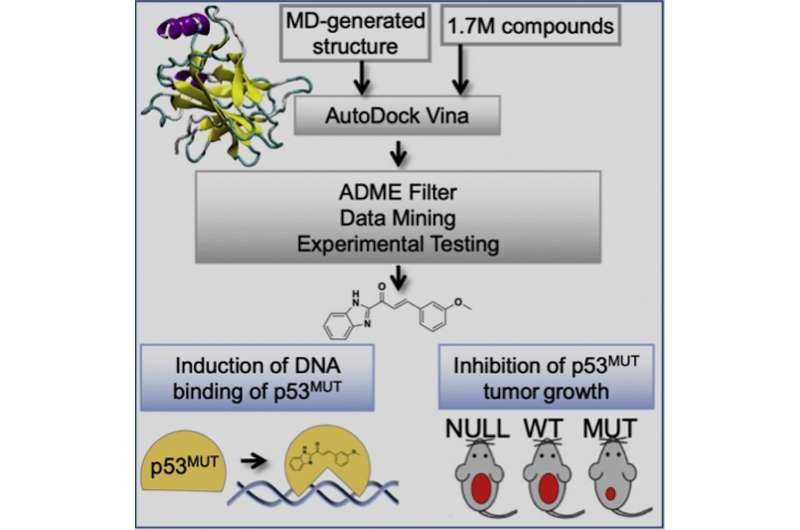
Published in Cell Chemical Biology, the study identified small drug-like compounds that act through a well-defined mode of action; do not require covalent attachment, induction of redox imbalance, or metal binding; and have selective anti-cancer activities on tumors with p53 missense mutations. This research provides a framework for p53 reactivation compound discovery that can help to increase chemical diversity and improve pharmacological properties necessary for translation of pharmaceutical p53 mutant reactivation to the clinic.
"This study successfully demonstrates the feasibility and efficacy of pharmaceutical reactivation of mutant p53," said Kaiser. "These findings are encouraging given the large number of cancer patients with p53 mutations that could benefit from such drugs."
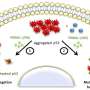
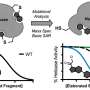
This study involved the application of an ensemble-based virtual screening approach, developed in the laboratory of Rommie Amaro, professor and endowed chair in the Department of Chemistry and Biochemistry at UC San Diego, which has the potential to identify compounds with increased cancer killing potential and with a broad spectrum of activity across a panel of p53 mutants. The researchers showed that their compounds bind mutant p53 and change mutant p53 conformation to wild type-like structures. This restores p53 DNA binding activity to activate the p53 transcriptional response, which in turn prevents tumor progression in mouse models selectively for tumors with a p53 missense mutation.
Challenges remain to define exact mechanisms and develop highly active corrector drugs for mutated p53 and future experiments are needed to optimize pharmacological properties to progress towards clinical therapeutics.
More information: Geetha Durairaj et al, Discovery of compounds that reactivate p53 mutants in vitro and in vivo, Cell Chemical Biology (2022). DOI: 10.1016/j.chembiol.2022.07.003
Journal information: Cell Chemical Biology
Provided by University of California, Irvine