Single-molecule experiments reveal force capability of artificial molecular motors
National University of Singapore physicists have shown that a single man-made molecular motor can exhibit a force similar to naturally occurring ones that power human muscles. Their results are published in Nanoscale.
Molecular motors are a class of machines with nanoscale dimensions that are essential agents of movement in living organisms. They harness various energy sources within the body to generate mechanical motion. A key characteristic is the force generated by a single motor during its self-propelled motion. This force generation capability allows the molecular motor to deliver mechanical work and is a measure of its energy conversion efficiency, which influences its use in potential applications.
The measurement of the force generated by naturally occurring molecular motors, which are usually made of proteins, was achieved two decades ago. However, similar measurements for artificial man-made molecular motors made of DNA (deoxyribonucleic acid) remain a challenge. A research collaboration between the Molecular Motors Laboratory by Associate Professor Zhisong Wang and the Single-Molecule Biophysics Laboratory by Professor Jie Yan, both from the Department of Physics, NUS has managed to detect the force generated by a moving DNA molecular motor.
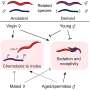
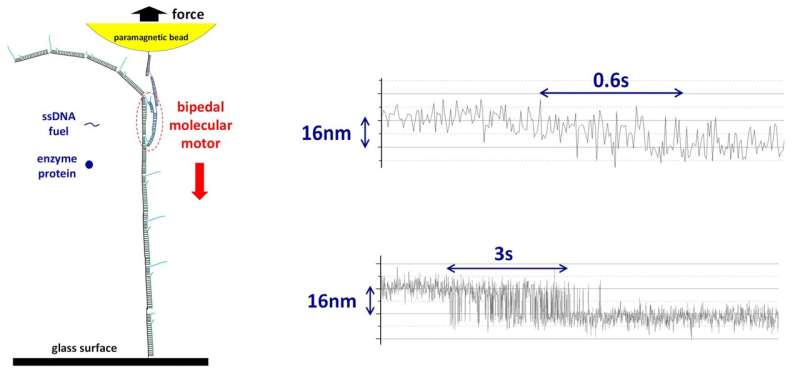
It is difficult to detect the forces created by a single molecular motor in motion for artificial motors because they are small and mostly operate on soft tracks (e.g., double-stranded DNA). Soft tracks are not fixed in position and tend to coil into a circular shape. This affects the motion of the artificial motor. The research team overcame this difficulty by designing and executing in parallel single-molecule experiments that kept the tracks in place at the nanoscale level while also simultaneously detecting the miniscule force created by the moving molecular motor. Using the magnetic tweezers technique, they first assembled an artificial molecular motor and its track under a paramagnetic bead (tool for isolation of biomolecules). They then switched the paramagnetic bead to a force detection mode (see Figure).
The research team successfully applied their method to an autonomous DNA molecular motor (previously developed by Prof. Wang's lab). This bipedal molecular motor is able to "walk" consecutively on its own with a stride length of about 16 nm between each step, providing a maximum force output of around 2 to 3 pN. This measured force output is at a level which is near to naturally occurring molecular motors powering human muscles, indicating a reasonably efficient conversion of chemical energy to mechanical motion.
Prof. Wang said, "This study paves the way for the development of applications associated with translational artificial molecular motors which require the generation of forces. Examples include molecular robots and biomimicking artificial muscles. Separately, the single-molecule method established in this work is applicable to force measurement of many other artificial molecular motors with soft tracks."
More information: Xinpeng Hu et al, Single-molecule mechanical study of an autonomous artificial translational molecular motor beyond bridge-burning design, Nanoscale (2021). DOI: 10.1039/D1NR02296B
Journal information: Nanoscale
Provided by National University of Singapore