Recent research on intramembrane proteases could lead to new Alzheimer's treatments
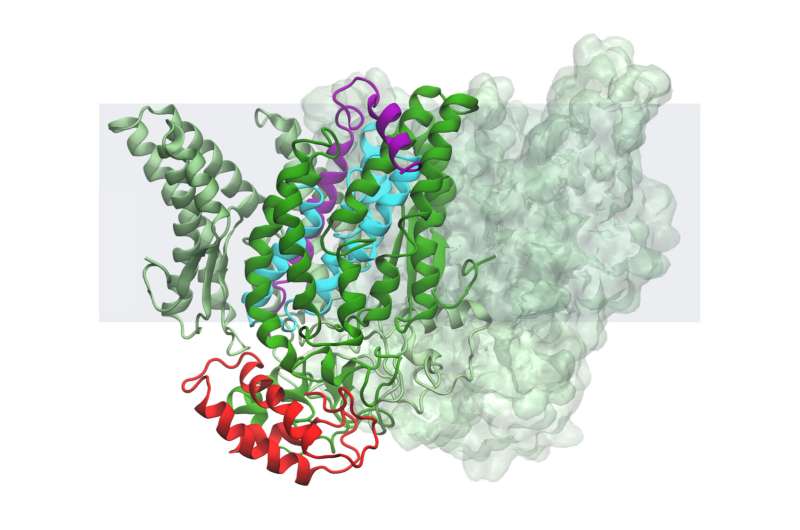
Working with tiny bacteria, Michigan State University researchers led by Lee Kroos have made a discovery that could have big implications for biology.
The researchers have revealed a new way that nature can inhibit or switch off important proteins known as intramembrane proteases, which the team reported April 26 in the journal eLife.
Although the researchers made this finding using a model organism, a microbe known as Bacillus subtilis, this type of protein is highly conserved, which is how evolutionary biologists say, "it's everywhere."
These types of proteases are found in organisms that span the kingdoms of life, from single-celled bacteria to people. In fact, the first intramembrane protease was discovered in humans in 1997 and perhaps the best-known member of this family, named gamma-secretase, is implicated in Alzheimer's disease.
"Our paper shows the first example of regulating an intramembrane protease with natural inhibitor proteins," said Kroos, a professor in the College of Natural Science's Department of Biochemistry and Molecular Biologyand Department of Microbiology and Molecular Genetics. "That gives us some ideas on how we might be able to use and mimic that.
"Will it tell us how to modulate gamma-secretase? No," Kroos said. "But it could give folks some ideas on decorations they could put on inhibitors to try as therapeutics."
Using this information to design drugs to treat Alzheimer's will take years, Kroos said, but the findings could have more immediate impacts in fighting particularly nasty and stubborn bacterial pathogens. That includes Bacillus anthracis, the bacterium behind anthrax infections, and other bacteria responsible for tetanus, botulism and food poisoning.
"Many, many bacteria have intramembrane proteases that are pretty closely related to the one we studied," said Kroos, who has been a fellow of the American Association for the Advancement of Science since 2014, thanks in part to his work furthering our understanding of biology with bacteria. "If we figure those out, we could figure out how to make bacteria less stress-resistant and more treatable with antibiotics."
A better understanding of these proteases could also help develop applications in other areas, including agriculture and environmental protection, eLife noted in a digest featuring the researchers' work. Beyond that, it helps paint a more complete picture of how life works.
"This work should have a broad impact influencing our understanding of the regulation of this class of proteins across the tree of life," wrote Petra Anne Levin, an editor for eLife and a professor of biology at Washington University in St. Louis.
Scissors, spores and Corvettes
A protease is an enzyme, a type of protein machine, that nature uses to chop up other proteins. This is a fundamental biological process that cells use to achieve a variety of goals. An intramembrane protease is an enzyme that has its active site—the spot where the enzyme does the snipping—buried inside a cell membrane.
"Sometimes you'll hear them called 'scissors in the membrane,'" Kroos said. "These intramembrane proteases do really important things in cells."
The protease studied by the researchers, for example, is part of the biological system that B. subtilis uses to make spores when food is scarce. Spores are essentially dormant cells coated in a protein armor that can weather harsh conditions and then reactivate once things improve (other bacteria including B. anthracis also form spores, which is one reason these pathogens are so persistent).
Because intramembrane proteases do their work within the confines of a cell membrane, it's been challenging for researchers to determine exactly how they work. Adding to the complexity of the project, the researchers thought their protease might be working in a sophisticated way that had never been documented before.
"When you look at other related organisms, you see that this system has evolved a lot," Kroos said. "B. subtilis is like the Corvette. It has the high-end machinery."
Understanding this high-end machinery required extensive genetic and biochemical testing, which was led by Sandra Olenic, a doctoral student in Kroos's lab. Olenic earned her Ph.D. after completing this project and is now a postdoctoral scholar at Tufts University.
While Olenic designed and ran experiments, she and Kroos realized their results wouldn't provide all the answers they sought. They turned to one of Kroos's longtime collaborators, Michael Feig, an MSU professor of biochemistry and molecular biology, to bring in computer modeling and help complete the expansive puzzle.
Lim Heo, a postdoctoral research associate in Feig's lab, had expertise in a computational technique that could predict protein structures. The technique has gained some broader attention recently thanks to Google and other big names in artificial intelligence developing software packages that make it more accessible to the scientific community.
Before such tools were available, however, Heo and Feig still had the know-how to help Kroos and Olenic start piecing together a model explaining how the intramembrane protease worked.
"I think this is a really cool story and a nice collaboration that took a lot of hard work and perseverance," Kroos said. "Sandra just had incredible persistence and dedication. It's a credit, too, to Lim and Michael that they did such a nice piece of work with the computational modeling."
Completing the puzzle
Taken altogether, the team's results imply that this B. subtilis intramembrane protease is kept inactive—that is, not snipping its target protein or substrate—with help from two other proteins. One of these inhibitor proteins works like a clamp, keeping the second protein lodged in the scissor enzyme's active site.
The researchers theorize that the bacteria can then activate the protease by releasing the clamp, letting the blocking protein slip out and allowing the target protein in.
"This was like putting together a 5,000-piece jigsaw puzzle without knowing what it looks like," Kroos said. Although the puzzle isn't completely solved, the team has enough data and results to be confident it has a reasonable model of how things look and work. But the researchers aren't stopping there.
Their next step is already in progress, which involves testing the predictions of the model—such as where the clamp is attached to both the protease and inhibitor protein—and seeing how well those match reality. Two undergraduate researchers in Kroos's lab are leading those experiments.
Another key step is determining the structures of the proteins involved using analytical techniques including X-ray crystallography and cryogenic electron microscopy. Kroos has been working on that part for about a decade with collaborators at MSU, but the proteins are in no hurry to give up their secrets.
Kroos suspects he may retire before the puzzle is totally finished, but that doesn't appear to bother him. For one, he confessed he might try to sneak back and help if his colleagues will let him. And he mostly seems excited to see how much of the puzzle he and his team can solve with the few years he has left.
When Kroos started at MSU in 1988, science didn't know about intramembrane proteases. But it wasn't long after their 1997 discovery that Kroos decided he needed to switch his research focus to include these proteins.
"That's not an easy thing to do," Kroos said, because not everyone—funding agencies included—believed the switch could be made. But he found support and willing collaborators at MSU.
"Our department is really strong in membrane proteins," he said. "I thought it would be a good chance to bloom where I was planted."
It's safe to say that hypothesis was confirmed.
More information: Sandra Olenic et al, Inhibitory proteins block substrate access by occupying the active site cleft of Bacillus subtilis intramembrane protease SpoIVFB, eLife (2022). DOI: 10.7554/eLife.74275
Journal information: eLife
Provided by Michigan State University