Patients' individual genomes may affect efficacy, safety of gene editing
Gene editing has begun to be tested in clinical trials, using CRISPR-Cas9 and other technologies to directly edit DNA inside people's cells, and multiple trials are recruiting or in the planning stages. A new study led by Boston Children's Hospital and the University of Montreal raises a note of caution, finding that person-to-person genetic differences may undercut the efficacy of the gene editing process or, in more rare cases, cause a potentially dangerous "off target" effect.
The study adds to evidence that gene editing may need to be adapted to each patient's genome, to ensure there aren't variants in DNA sequence in or near the gene being targeted that would throw off the technology. Findings appear this week in the Proceedings of the National Academy of Sciences (December 11-15).
"Humans vary in their DNA sequences, and what is taken as the 'normal' DNA sequence for reference cannot account for all these differences," says Stuart Orkin, MD, of Dana-Farber Boston Children's Cancer and Blood Disorders Center and co-corresponding author on the study with Matthew Canver, an MD-PhD student at Harvard Medical School. "We recommend that common variation be taken into account in designing targeting systems for therapeutic editing, to maximize efficacy and minimize potential safety concerns."
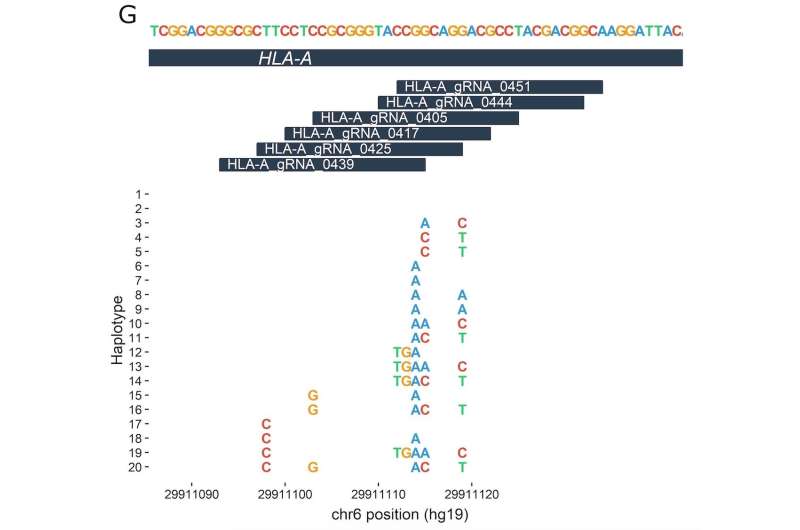
The study analyzed 7,444 previously published whole-genome sequences. Based on a list of about 30 disease-related DNA targets that researchers are interested in altering through gene editing, the researchers made a second list of nearly 3,000 guide RNAs (gRNAs). These are bits of genetic code that have been developed to direct CRISPR-Cas9 enzymes to the right editing location on or adjacent to the target, like the address on an envelope.
The team, led by Orkin, Canver and Samuel Lessard of the University of Montreal, then looked to see whether any of the 7,444 individuals carried DNA sequence variants ("letter changes" or insertions/deletions) in the areas the gRNAs are looking for.
"If there are genetic differences at the site that CRISPR reagents are targeting for therapy, you are at risk for decreased efficacy or treatment failure," explains Canver, who conceived and led the study in Orkin's Boston Children's Hospital lab. "A difference in just a single base pair can cause a decrease in binding efficiency due to a mismatch with the guide RNA. Overall, this can cause a reduction in treatment efficacy."
The team found that such occurrences in the genome are not uncommon; about 50 percent of the analyzed gRNAs had the potential to be affected by variants at their target sites. In a few cases, the team found genetic variants that cause DNA sequences in the genome to more closely match a gRNA that could potentially draw it to the wrong place—resulting in an edit of a gene or other DNA region that's not meant to be targeted.
"In rare cases, there was the potential to create very potent 'off-target' sites - where CRISPR reagents could bind and cut where they're not intended to," says Canver. "If an off-target effect happens to be in, say, a tumor suppressor gene, that would be a big concern."
Although the study looked at CRISPR-Cas9 gene editing, the researchers believe their findings extend to other gene-editing tools such as zinc-finger nucleases (ZFN) and TAL effector nucleases.
"The unifying theme is that all these technologies rely on identifying stretches of DNA bases very specifically," says Canver. "So, a variant that affects the target sequence could reduce guide RNA binding. Variants can also lead to binding at new sites that could potentially cause harm. As these gene-editing therapies continue to develop and start to approach the clinic, it's important to make sure each therapy is going to be tailored to the patient that's going to be treated."
More information: Samuel Lessard el al., "Human genetic variation alters CRISPR-Cas9 on- and off-targeting specificity at therapeutically implicated loci," PNAS (2017). www.pnas.org/cgi/doi/10.1073/pnas.1714640114
Journal information: Proceedings of the National Academy of Sciences
Provided by Children's Hospital Boston